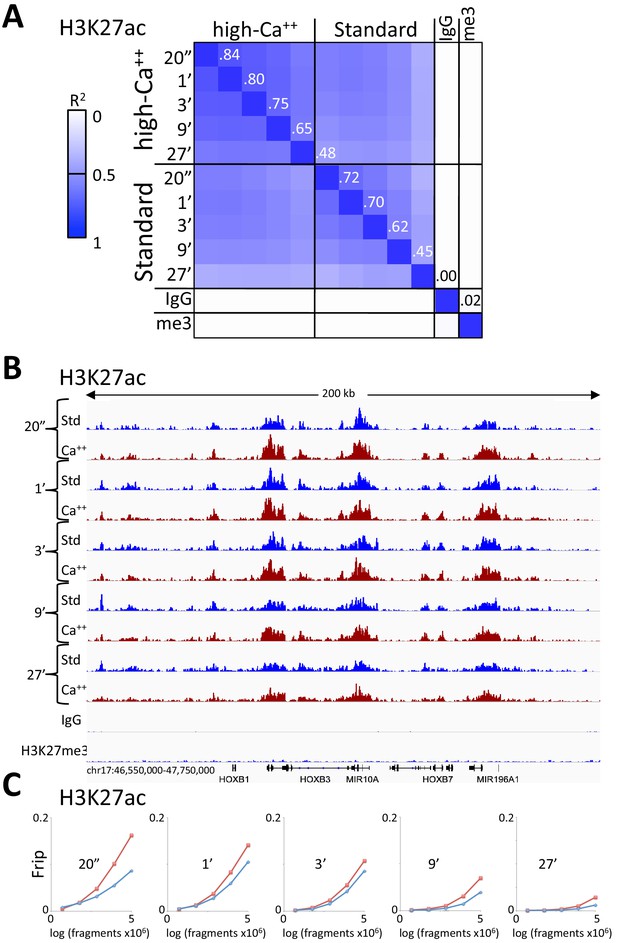
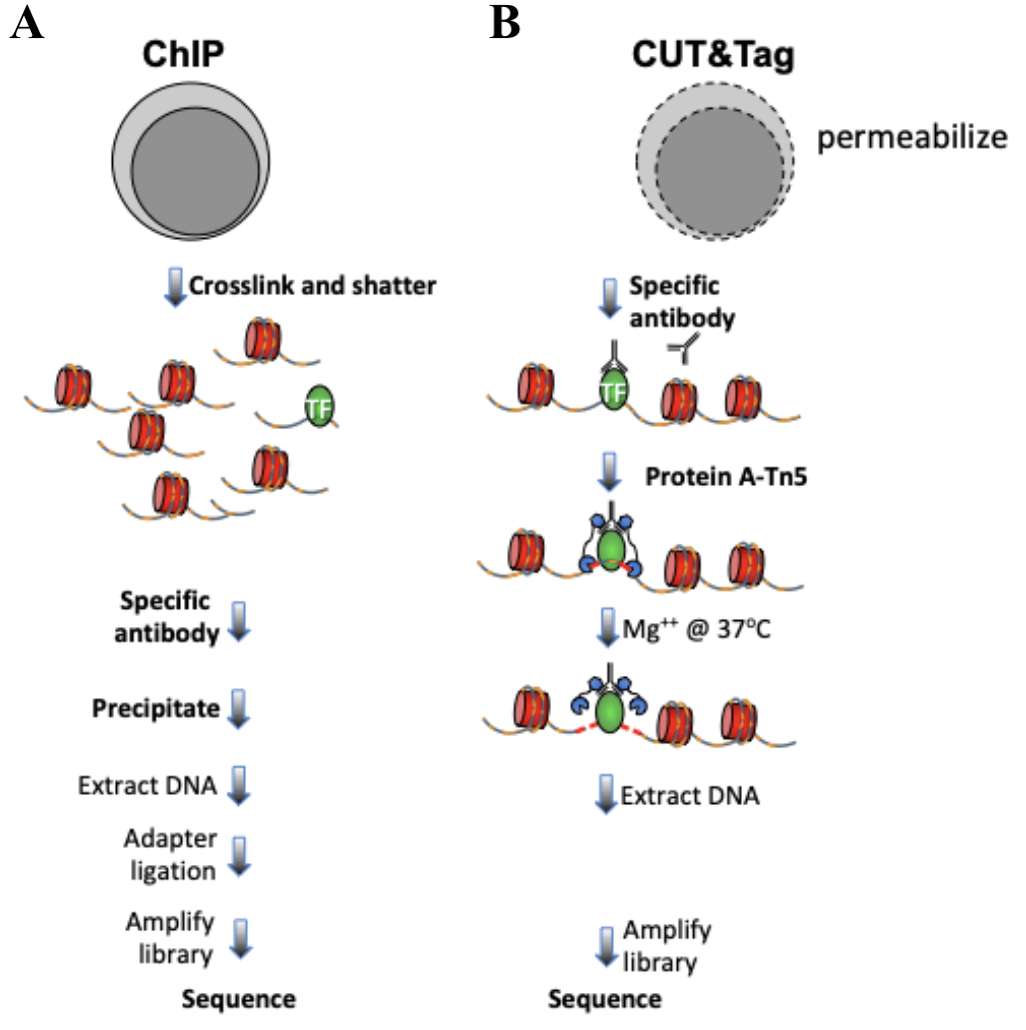
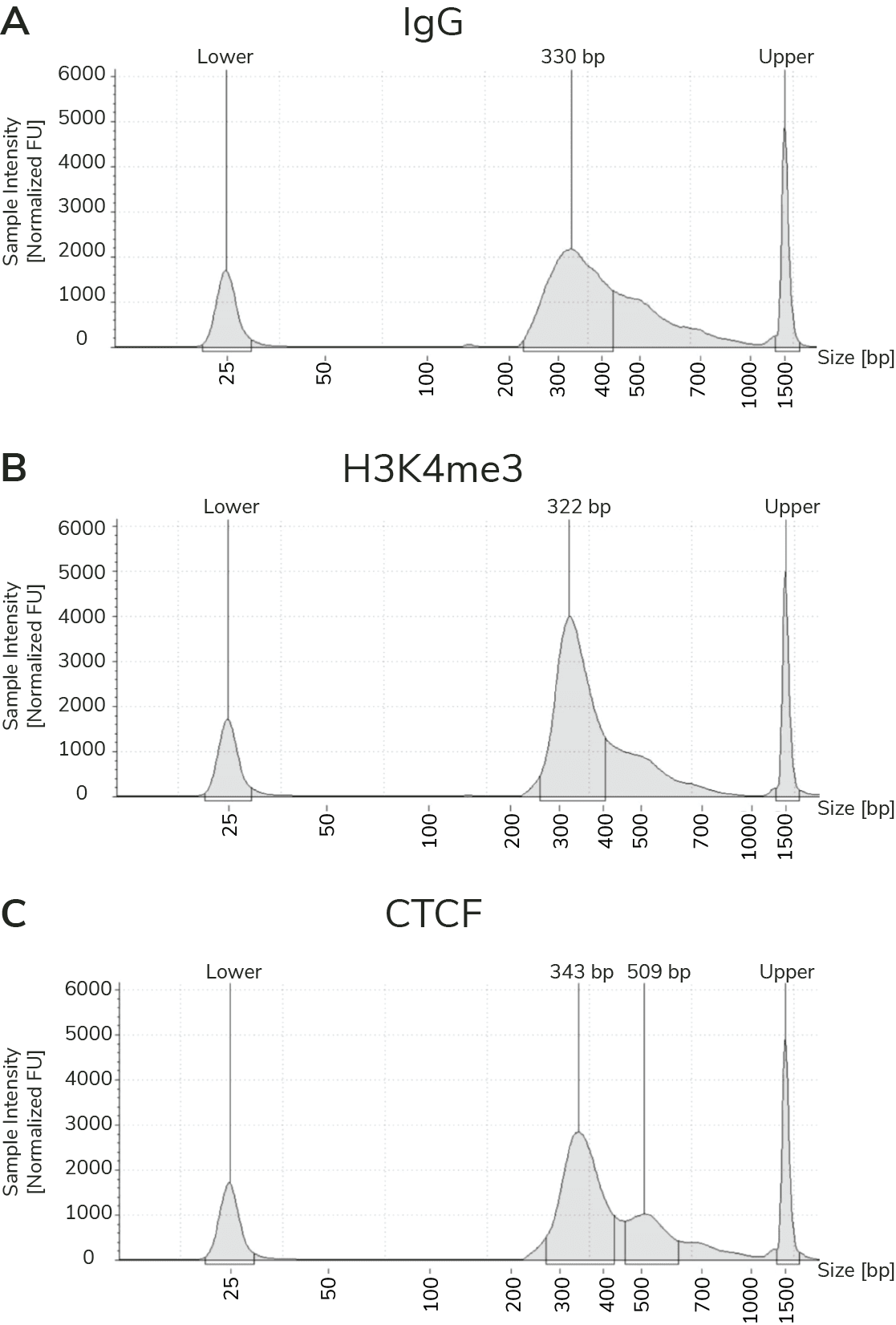
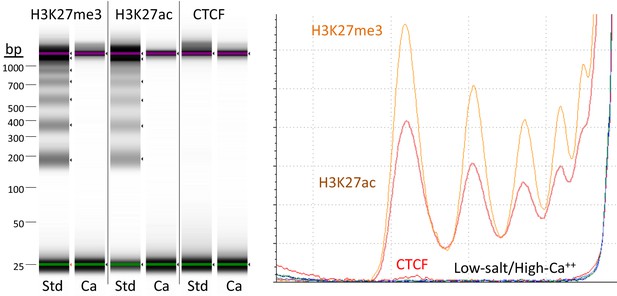
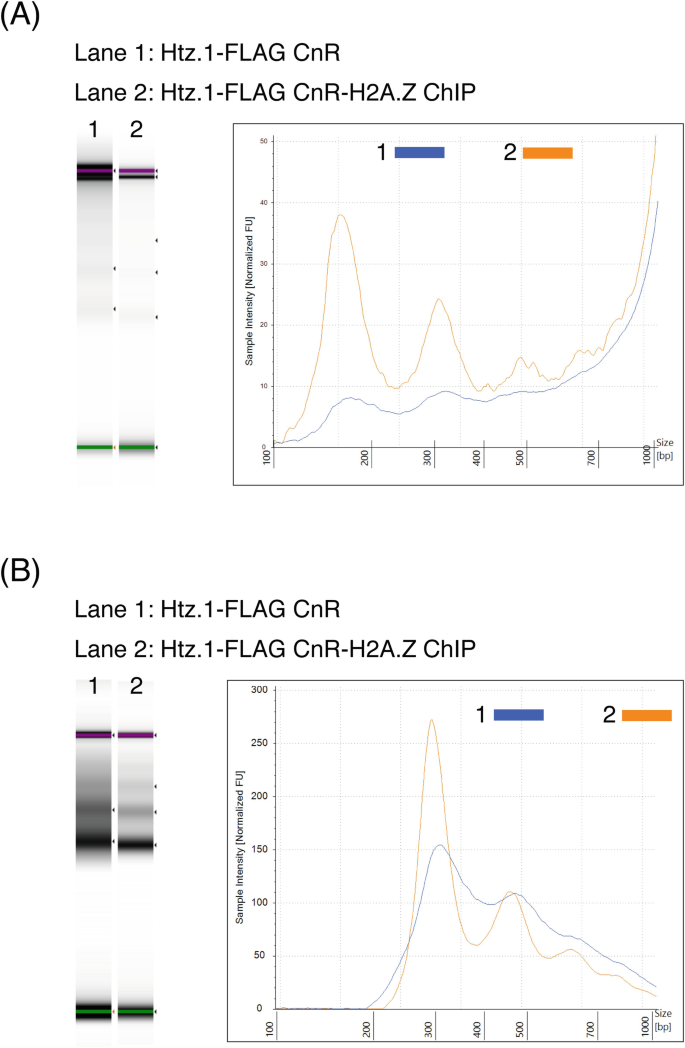
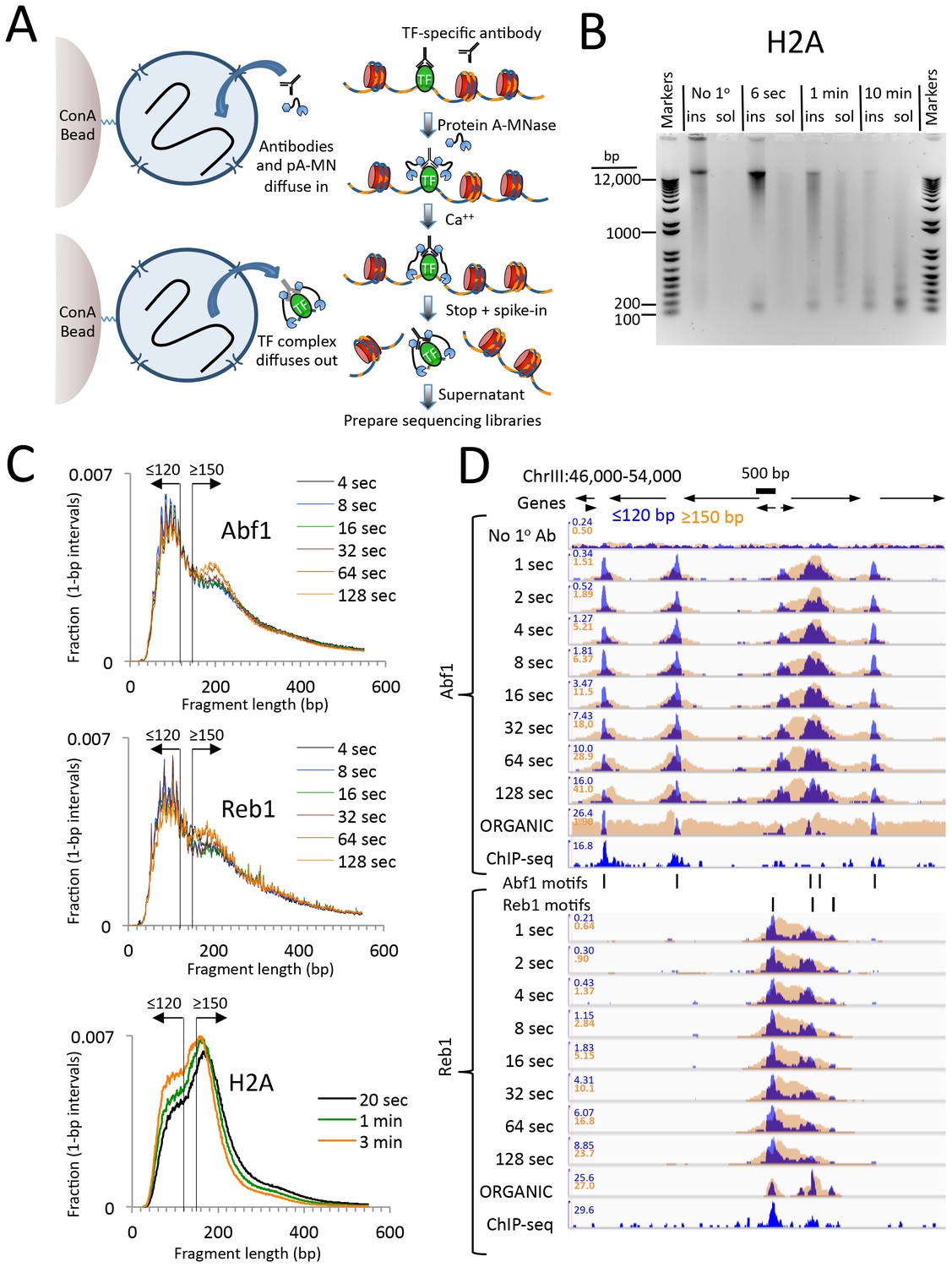
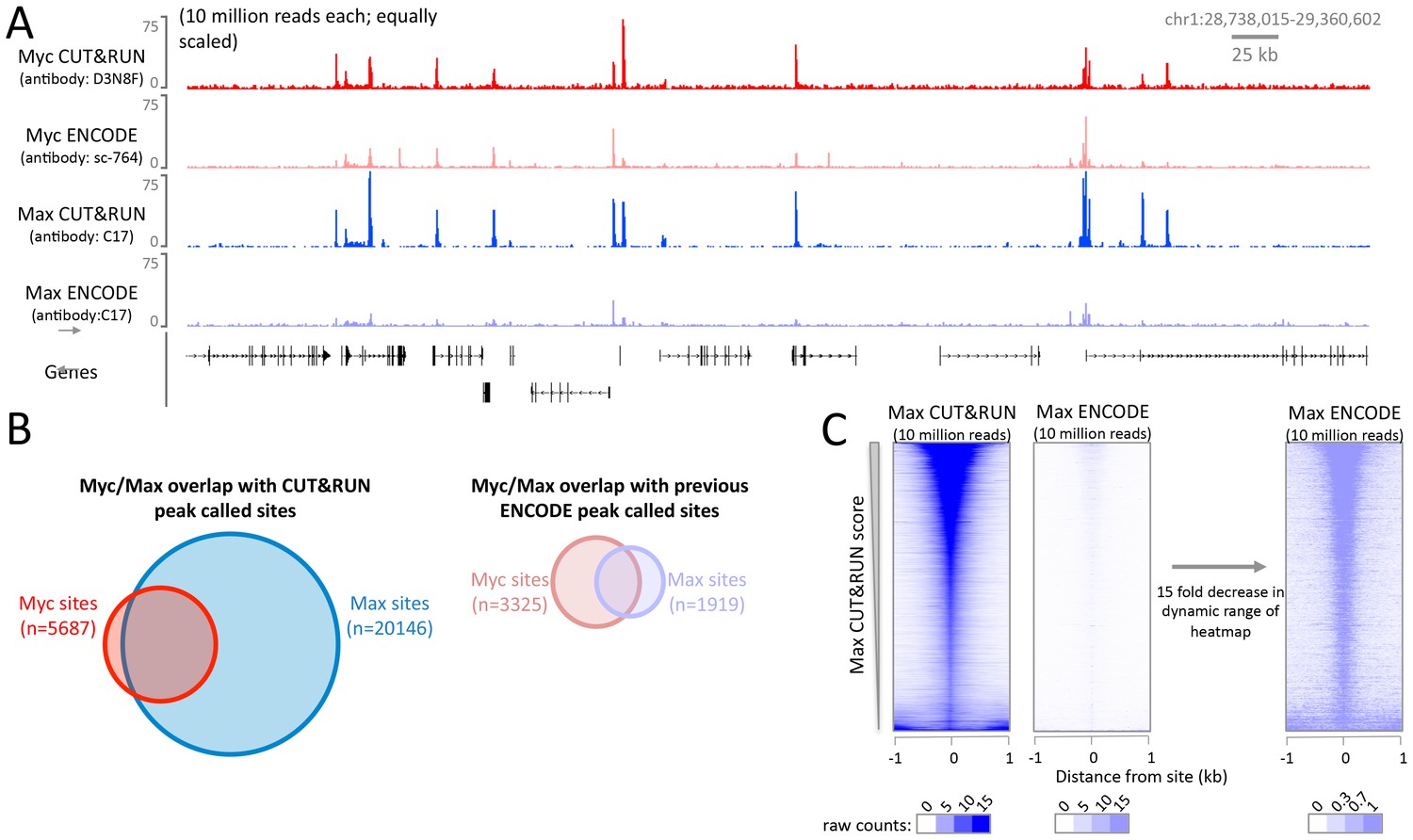
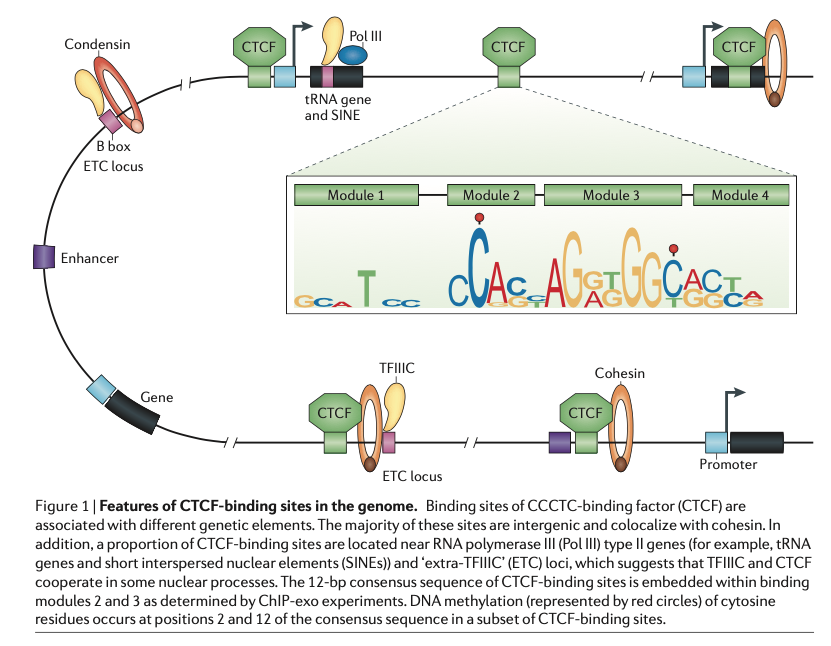
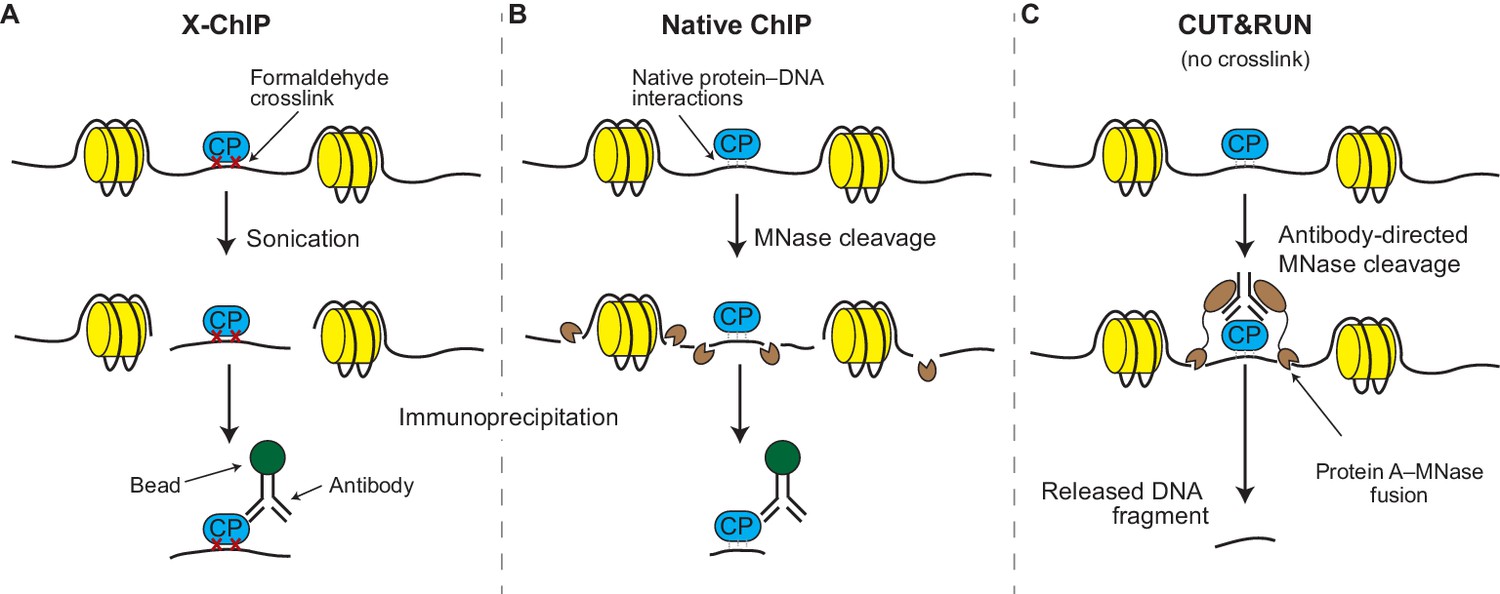
A modified CUT&RUN-seq technique for qPCR analysis of chromatin-protein interactions - ScienceDirect
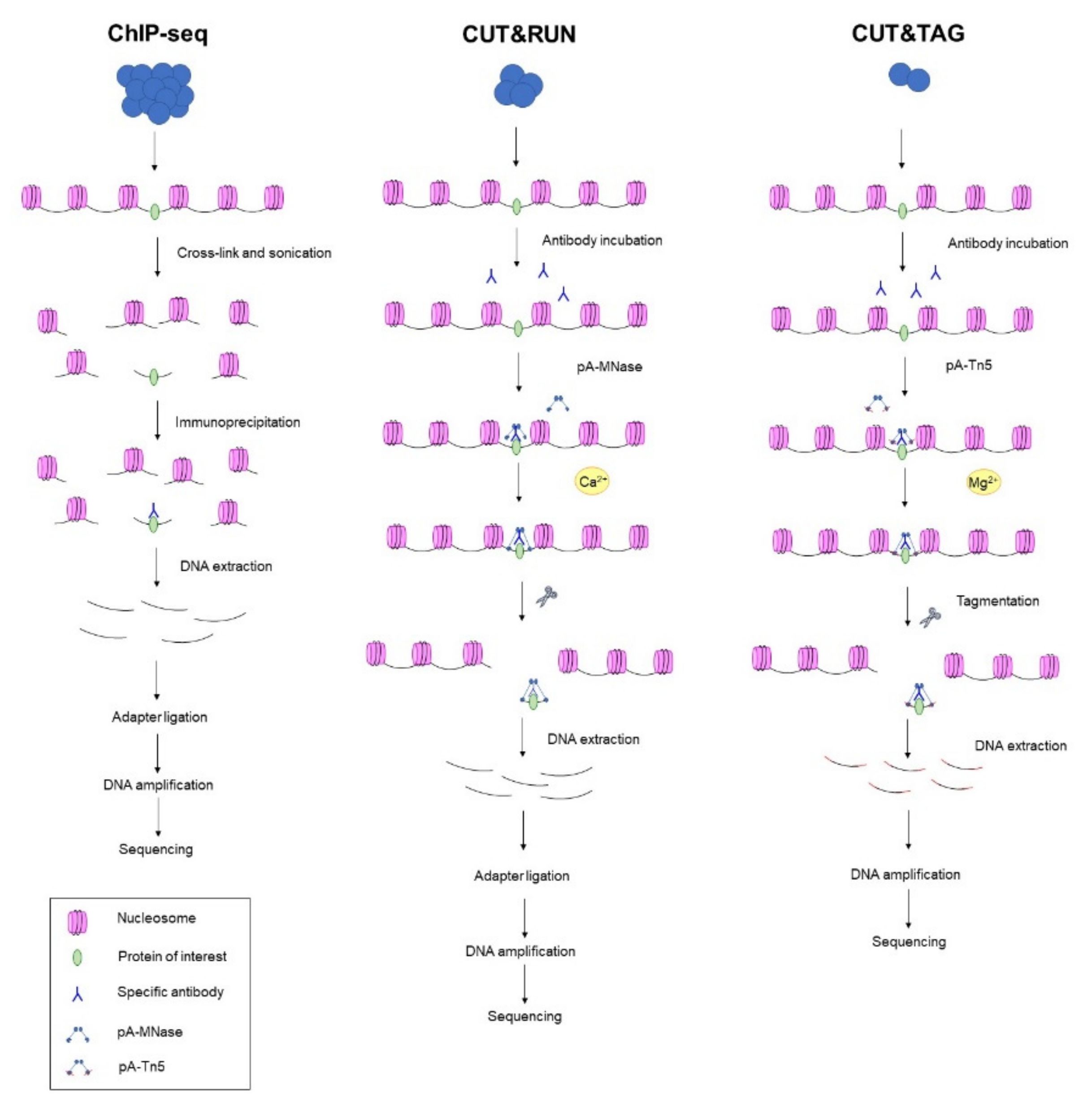
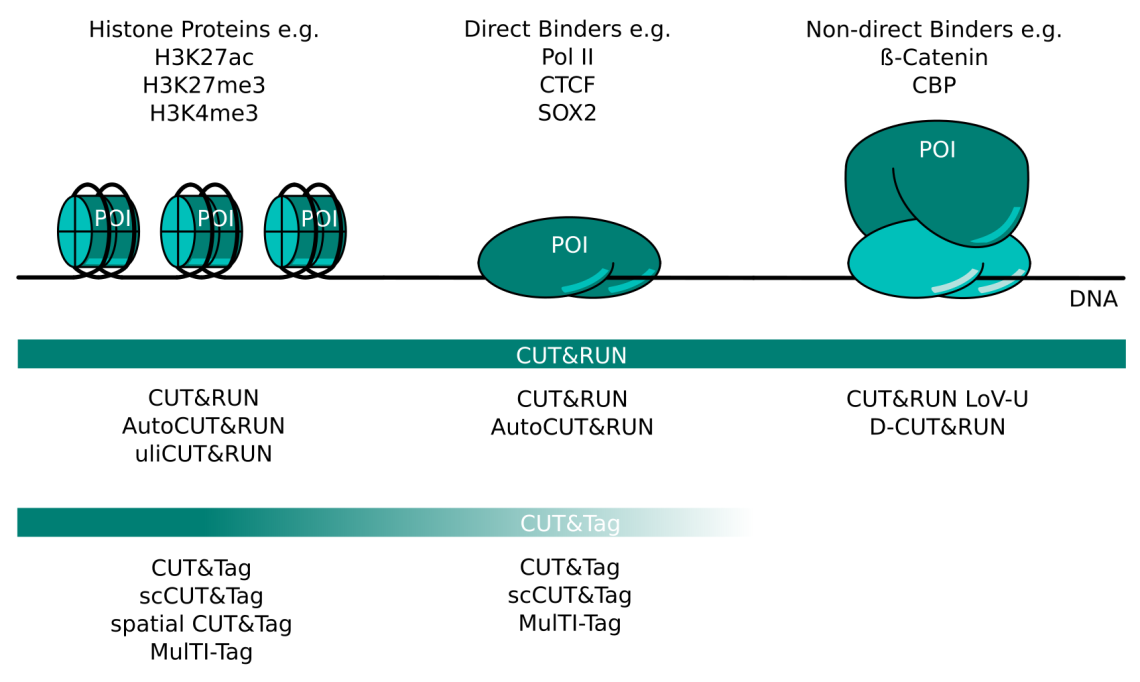
IJMS | Free Full-Text | Emerging Single-Cell Technological Approaches to Investigate Chromatin Dynamics and Centromere Regulation in Human Health and Disease
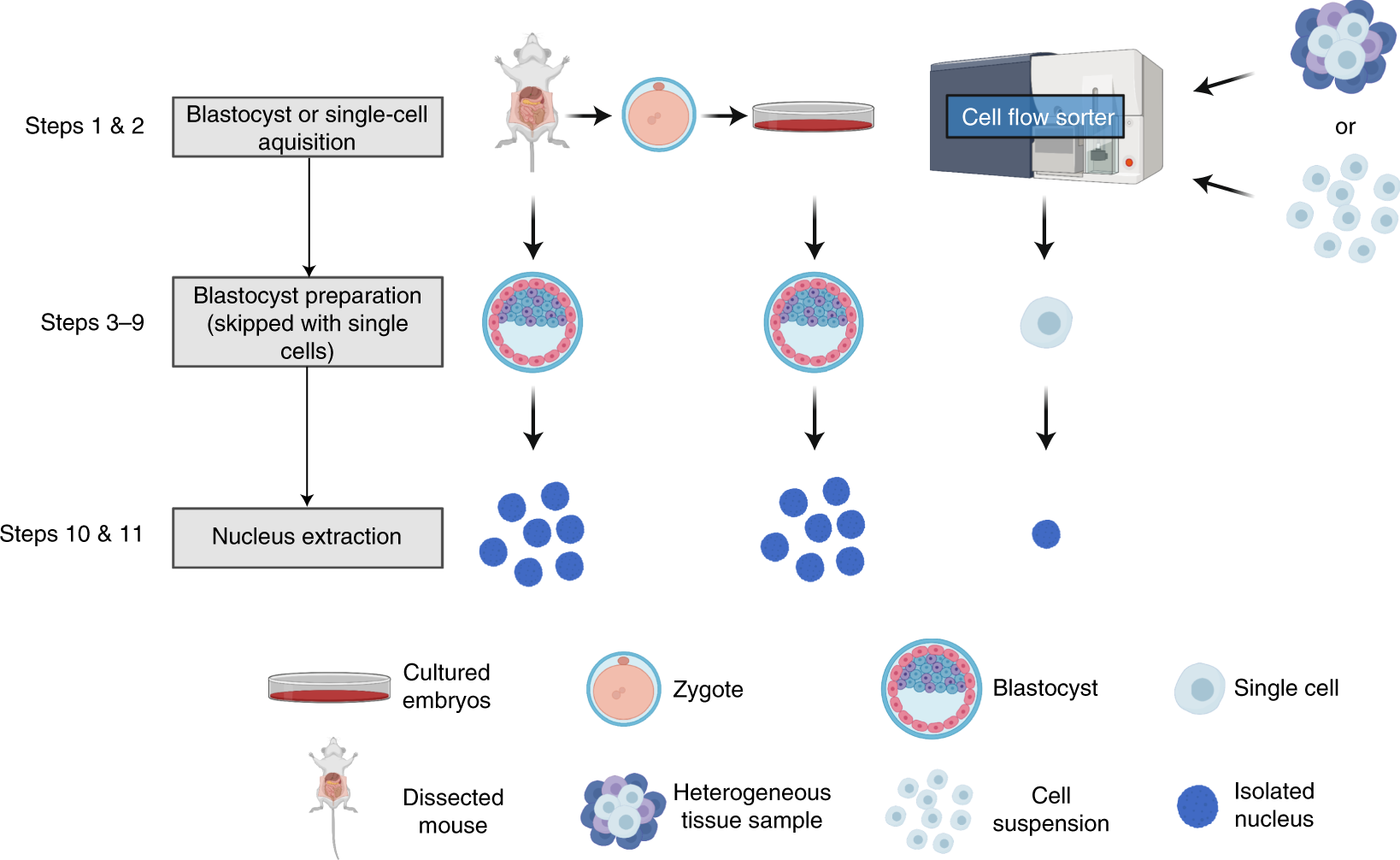
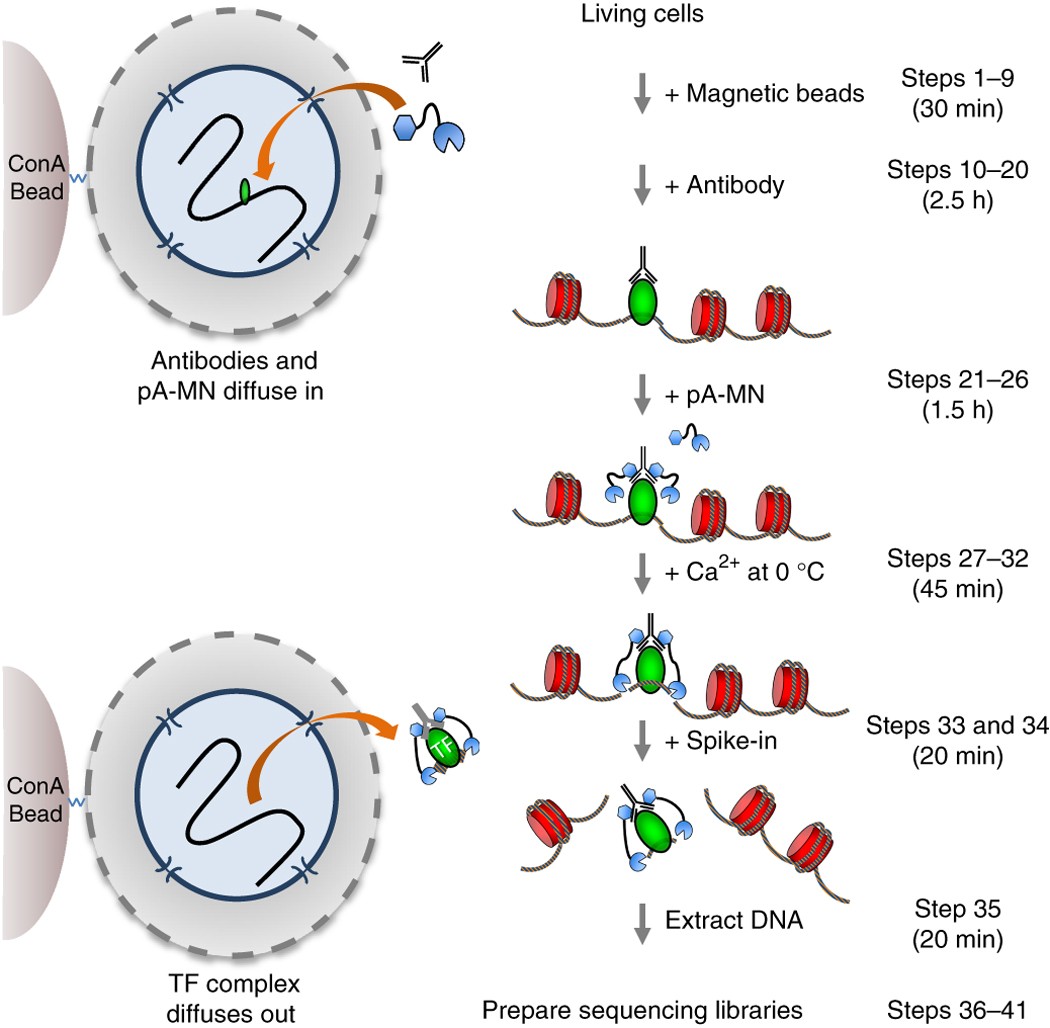
Transcription factor chromatin profiling genome-wide using uliCUT&RUN in single cells and individual blastocysts | Nature Protocols