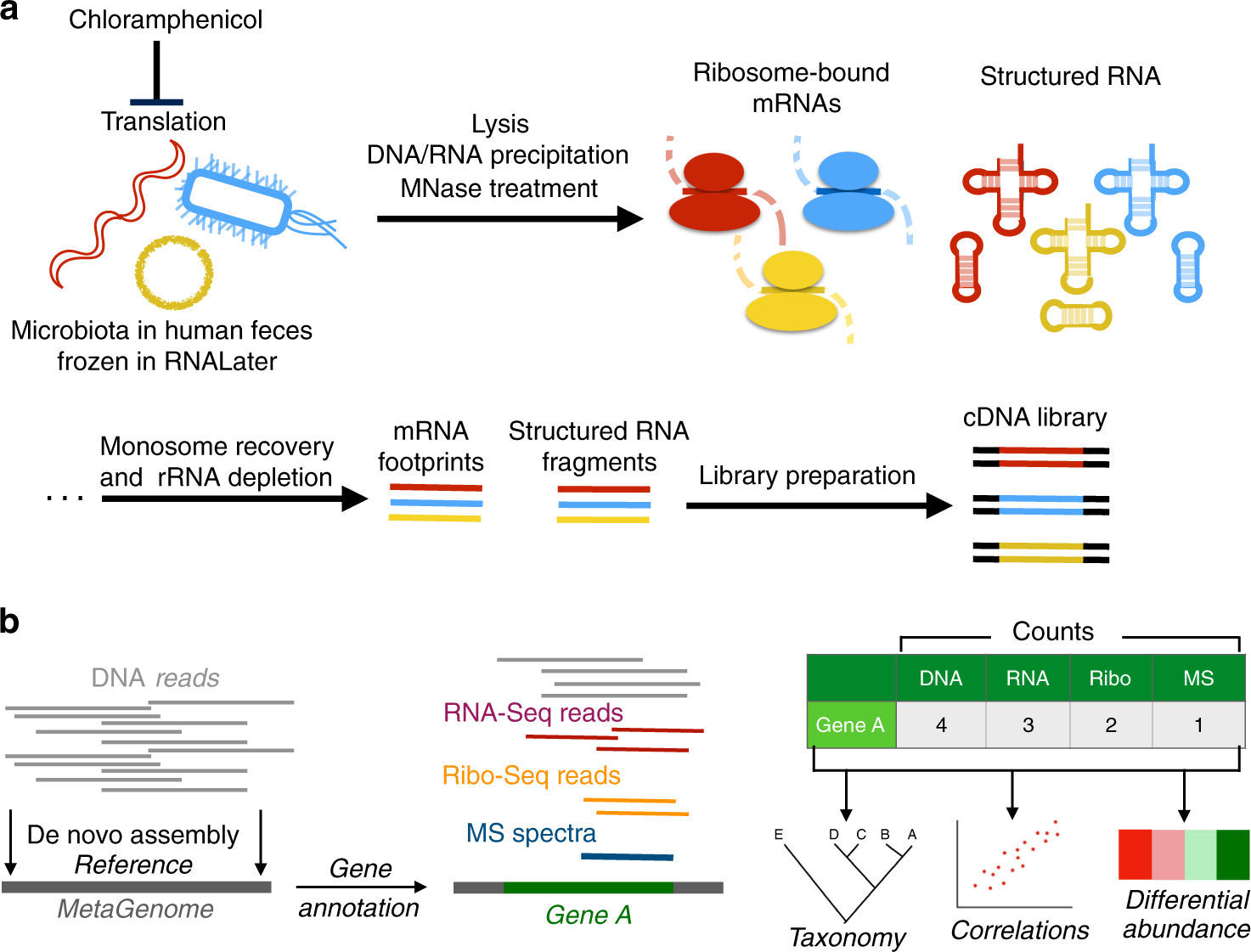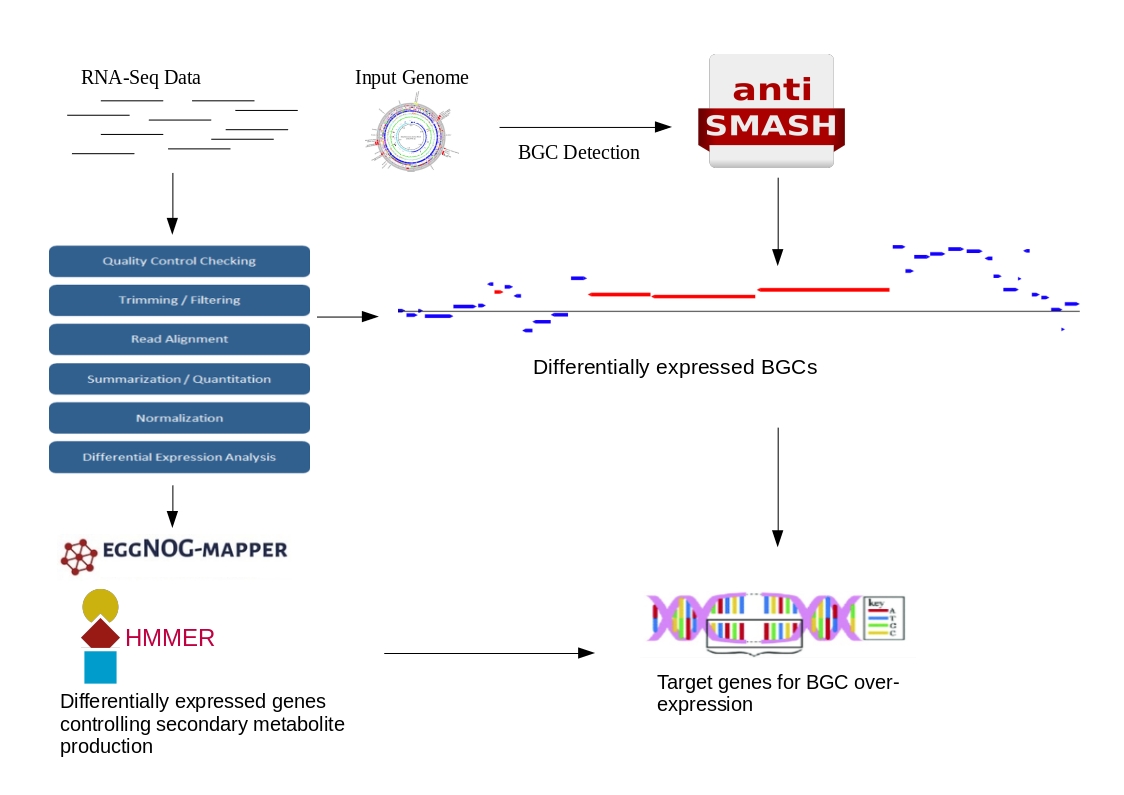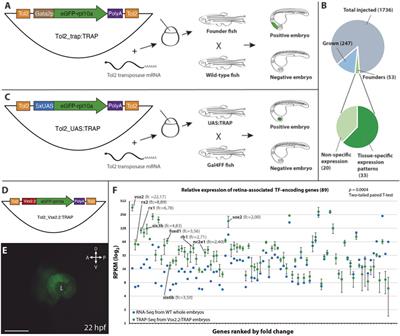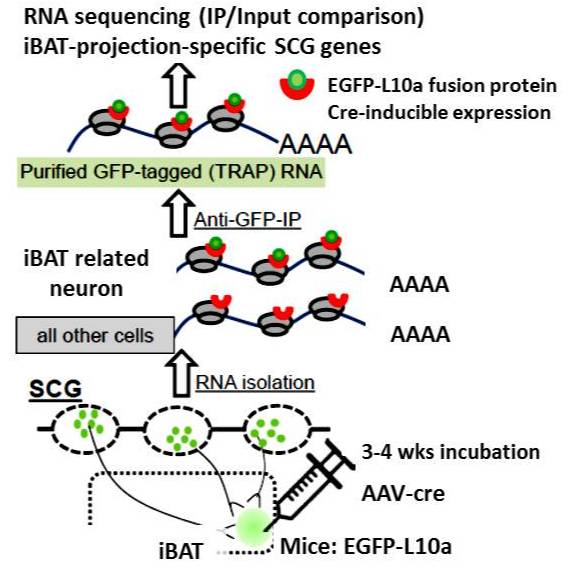
TRAP-SEQ (Translating Ribosome Affinity Purification followed by RNA sequencing) of interscapular brown adipose tissue (iBAT)- related ganglia from 7-day cold and warm treated mice - Blackfynn Discover

Translational changes induced by acute sleep deprivation uncovered by TRAP- Seq | Molecular Brain | Full Text

TRAP-SEQ of Eukaryotic Translatomes Applied to the Detection of Polysome-Associated Long Noncoding RNAs | SpringerLink
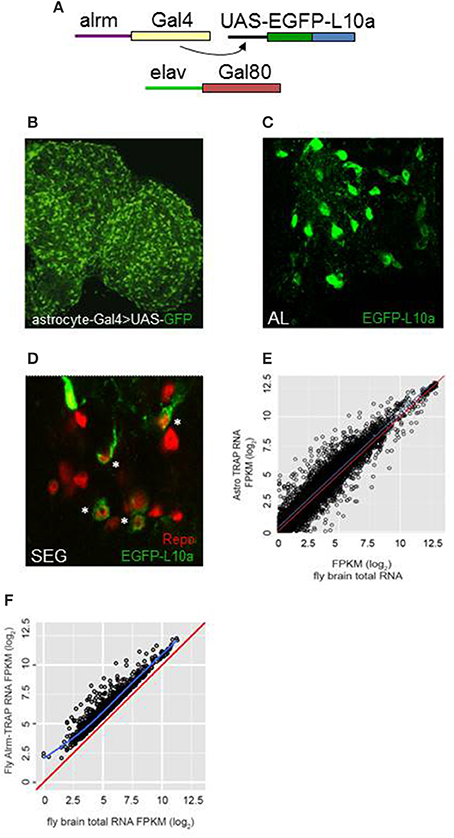
Frontiers | TRAP-seq Profiling and RNAi-Based Genetic Screens Identify Conserved Glial Genes Required for Adult Drosophila Behavior

Cells | Free Full-Text | Tracing Translational Footprint by Ribo-Seq: Principle, Workflow, and Applications to Understand the Mechanism of Human Diseases
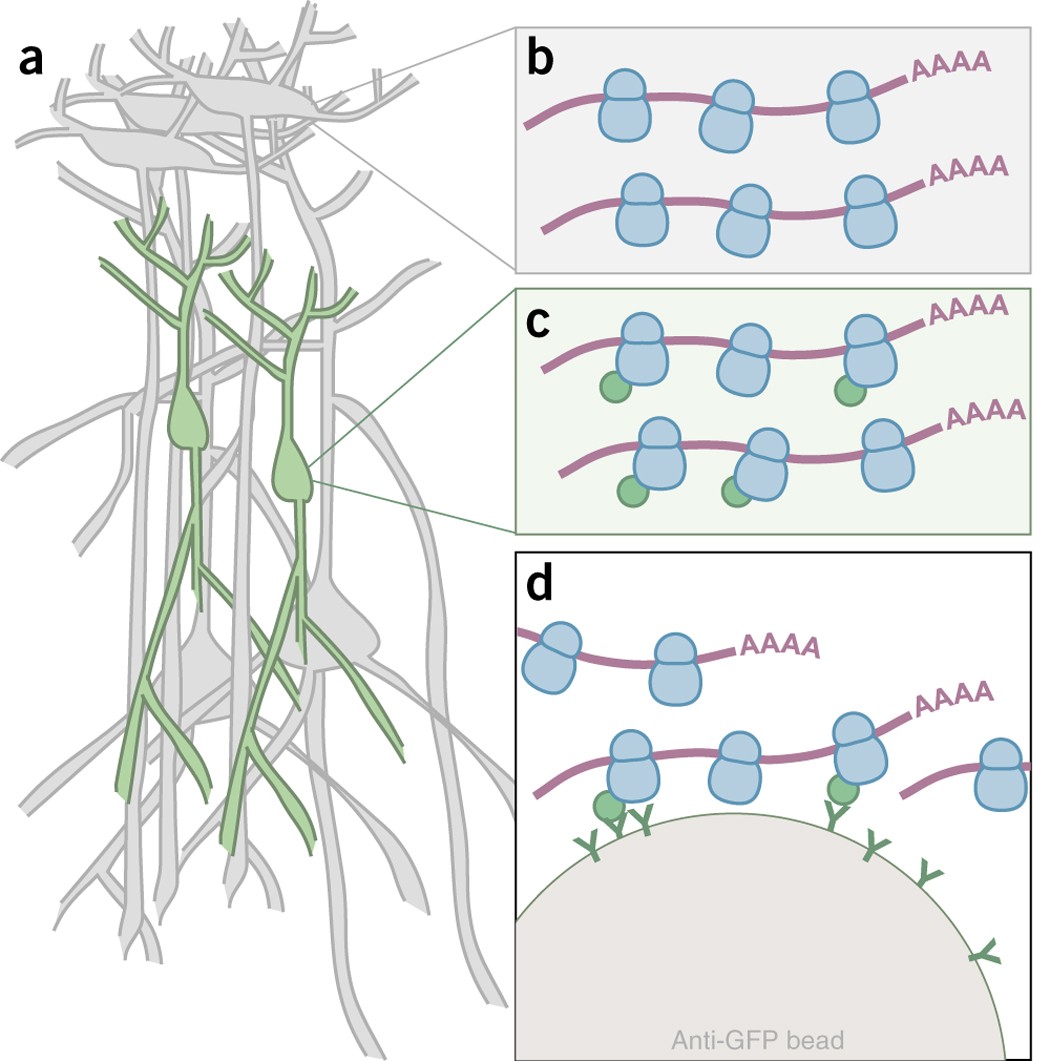
Cell type–specific mRNA purification by translating ribosome affinity purification (TRAP) | Nature Protocols

Translating Ribosome Affinity Purification (TRAP) Followed by RNA Sequencing Technology (TRAP-SEQ) for Quantitative Assessment of Plant Translatomes | SpringerLink

Translational changes induced by acute sleep deprivation uncovered by TRAP- Seq | Molecular Brain | Full Text
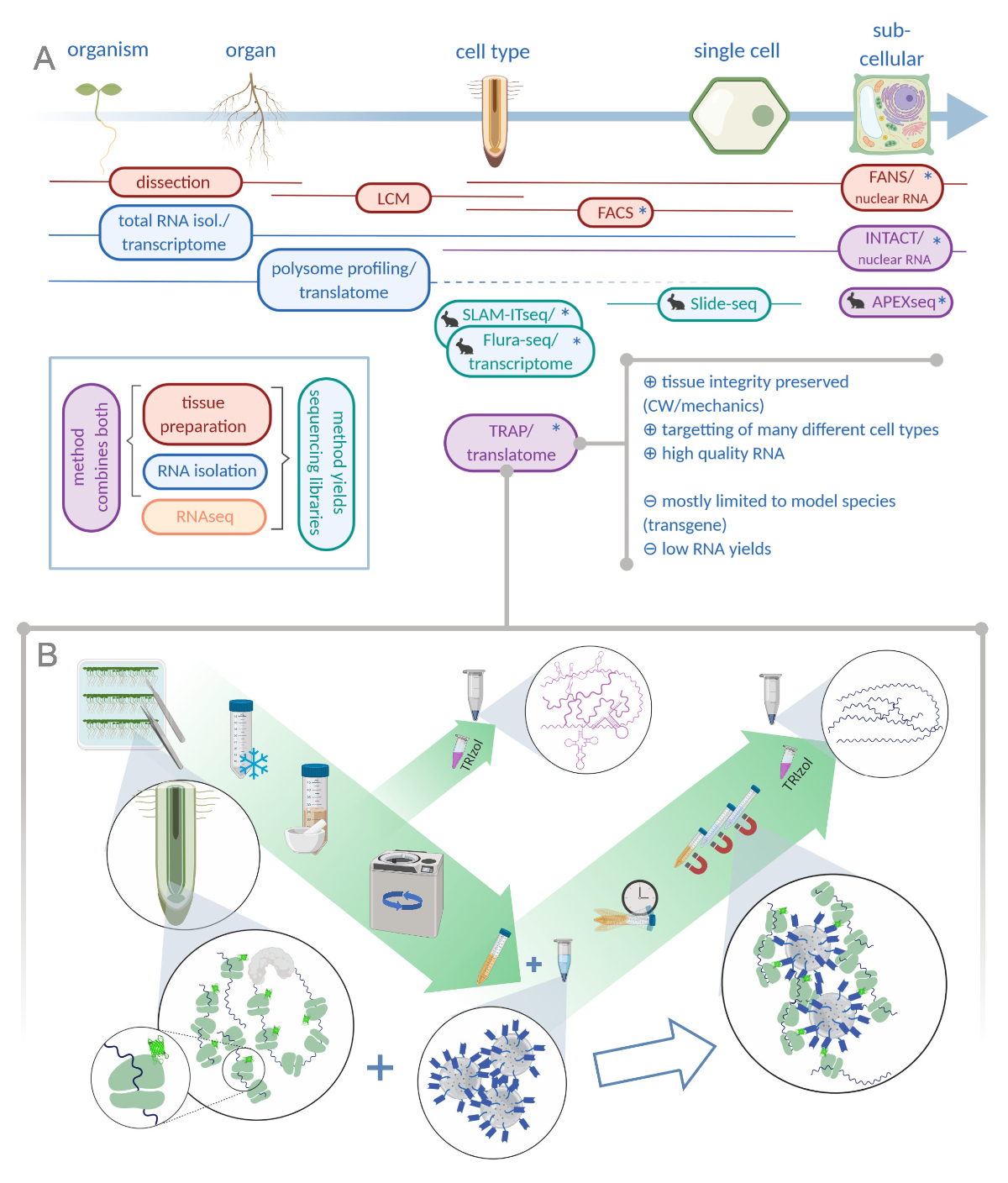
Translating Ribosome Affinity Purification (TRAP) to Investigate Arabidopsis thaliana Root Development at a Cell Type-Specific Scale | Protocol

Translational changes induced by acute sleep deprivation uncovered by TRAP- Seq | Molecular Brain | Full Text

Massively parallel quantification of CRISPR editing in cells by TRAP-seq enables better design of Cas9, ABE, CBE gRNAs of high efficiency and accuracy | bioRxiv
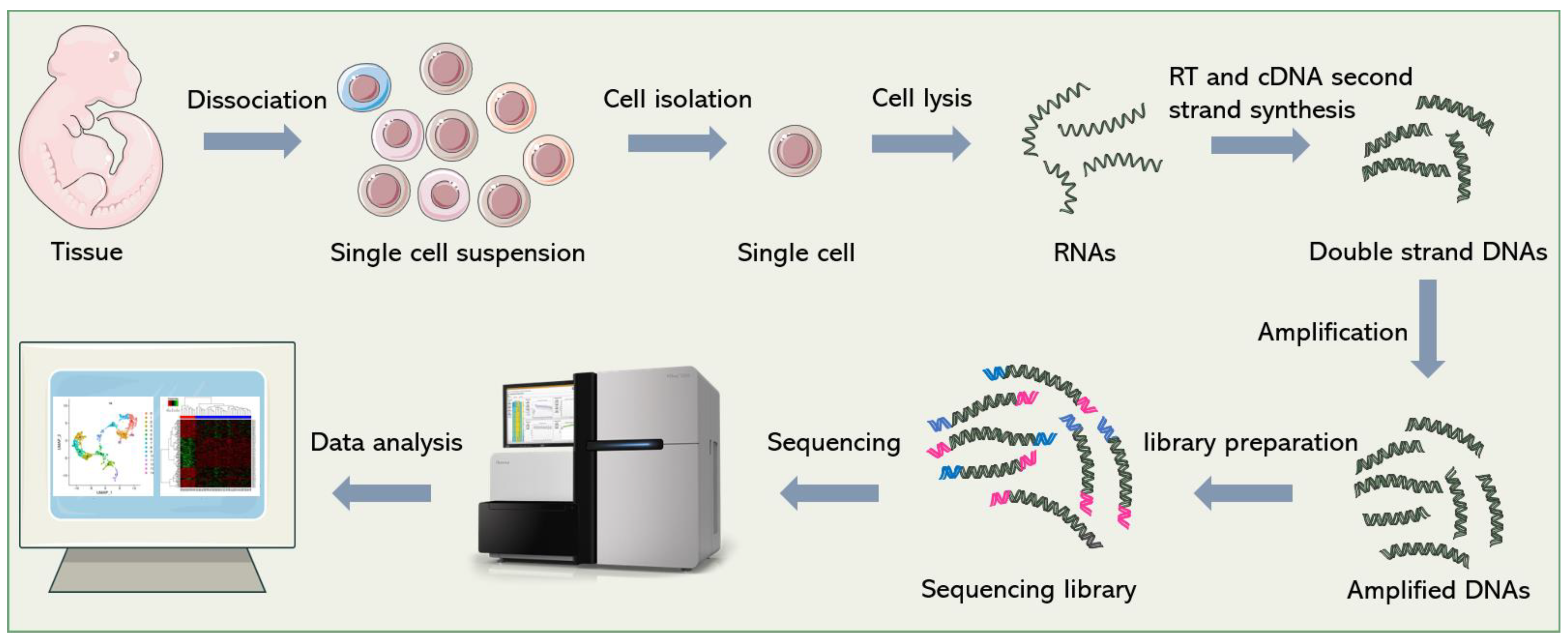
Biosensors | Free Full-Text | Microfluidics Facilitates the Development of Single-Cell RNA Sequencing

Massively parallel quantification of CRISPR editing in cells by TRAP-seq enables better design of Cas9, ABE, CBE gRNAs of high efficiency and accuracy | bioRxiv